Prof.Feng Chen’s Lab, School of Public Health, Nanjing MedicalUniversity
Gliomasaccount for 80% of primary intracranial malignant tumors. Low-gradegliomas (LGG) are characterized by wide heterogeneity, difficulttreatment and poor prognosis, which seriously endanger the physicaland mental health of human. Accurate prediction for patientprognosis, identification of patients at high mortality risk, andfurther individualized treatment are the cores of precision medicine,which has important values for public health and clinical practice.However, virtually few accurate and robust prediction models existfor LGG prognosis that may aid physicians in making clinicaldecisions.
Recently,with the in-depth collaboration between the Labs of Professor FengChen from the School of Public Health, Nanjing Medical University,Professor David C. Christiani from Harvard T.H. Chan School of PublicHealth, and Professor Yi Li from the School of Public Health,University of Michigan, by leveraging multiple prospective gliomacohorts around the world, and utilizing novel data mining andmodeling strategies, they developed an accurate and independentlyvalidated prediction model of lower-grade gliomas overall survival(APOLLO), which published as a Research Article in eBioMedicine(IF=11.205, Chinese Academy of Sciences /JCR Q1), an internationalauthoritative journal subordinate toTheLancet Discovery Science.

Thestudy was carried out according to the process of "BiomarkerScreening → Model Construction → Systematic Assessment"(Figure1).
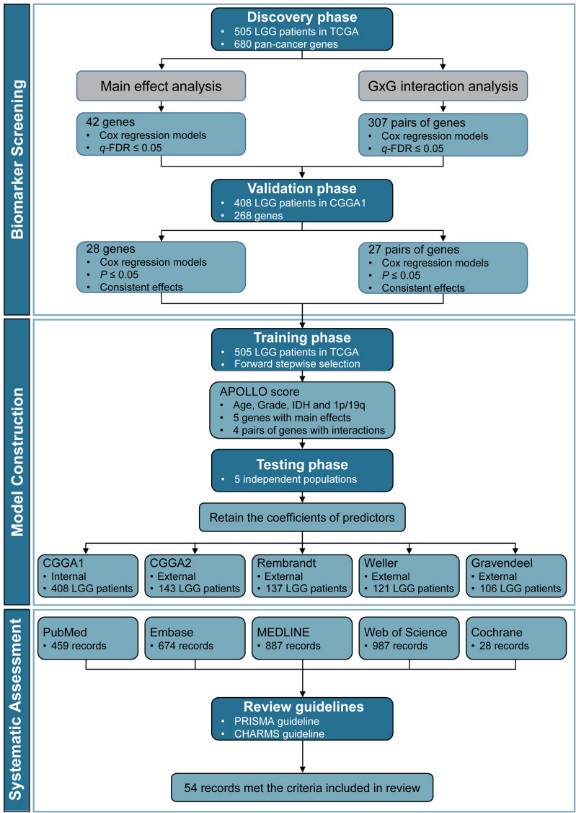
Figure1. Flowchart of development and validation of APOLLO and a systematicassessment
Todeal with the statistical challenges in high-dimensional datamodeling, improve the accuracy and robustness of prediction models,Professor Feng Chen's Lab innovatively proposed and applied a 3-Danalysis strategy, involving DoubleTypes of Effects (Identifyingbiomarkers with main effects or interaction effectssimultaneously,which ensured the accuracy), DoubleSteps of Screening(Screening biomarkers in the discovery set with correction ofmultiple tests, then validating biomarkers in an independentpopulation, which guaranteed the robustness of screened biomarkers)and DoubleSteps of Modeling (ConstructingAPOLLO in the training set, then retaining the coefficients ofpredictors and evaluating the performance in 1 internal testing setand 4 external testing set, which guaranteed the robustness ofAPOLLO).
APOLLOhad an adequate discriminative ability for mortality risk in all sixcohorts (Figure2 A-F).Further, patients were classified into six risk classes according tothe quintiles and 90% percentile of the APOLLO risk score. Patientswith APOLLO score >90% percentile had a 54.18 times of mortalityrisk than those with APOLLO score <20% (Figure2 G-H),suggesting APOLLO has the ability to identify patients at highmortality risk.
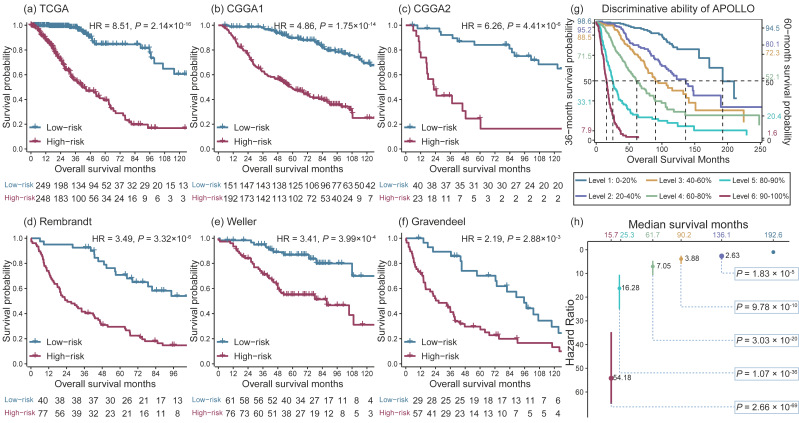
Figure2. Kaplan-Meier survival curves of LGG patients stratified by APOLLOscore.
APOLLOexhibited an excellent predictive ability for the 36- and 60-monthsurvival for LGG patients in all six independent cohorts (Figure3 A-F),Overall, the prediction accuracywas0.901 (0.879-0.923) for 36-month survival, and 0.843 (0.815-0.871)for 60-month survival, respectively. Additionally, APOLLO presentedan excellent C-index in six cohorts, with a pooled C-index of 0.818(95%CI: 0.800-0.835) (Figure3 G-I),indicating that the prediction accuracy of APOLLO was satisfactoryand robust.
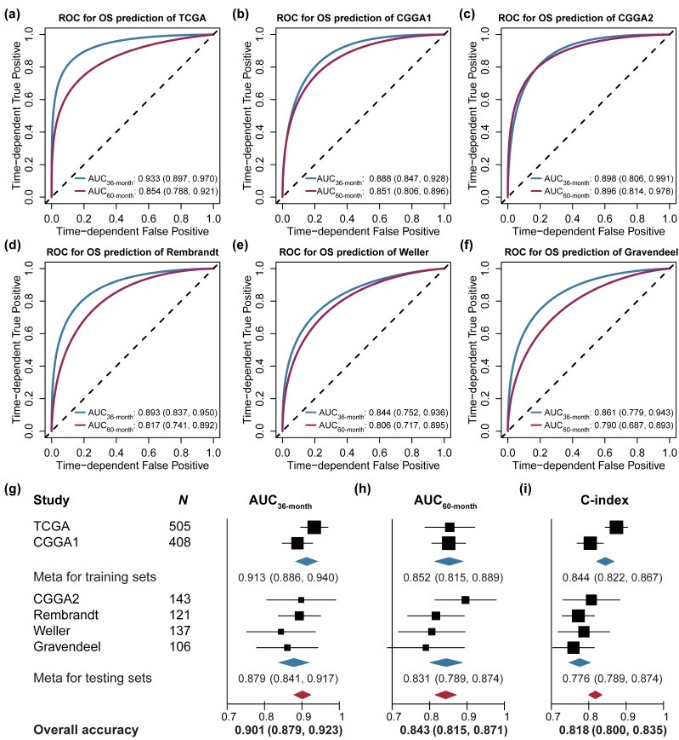
Figure3. Time-dependent receiver operating characteristic curves of APLLOfor 36- and 60-month overall survival prediction.
Followingthe PreferredReportingItemsfor SystematicReviewsand Meta-Analyses(PRISMA), Professor Feng Chen's Lab conducted the first systematicreview on prognostic prediction models of LGG. Compared with theexisting models, APOLLO has the largest sample size, the largestnumber of validation sets, and the most accurate and robustperformance for prognostic prediction of LGG.
Decisioncurve analysis (DCA) showed that APOLLO presented more clinical netbenefits than several competing intervention strategies (Figure4 A-D).According to Global Burden of Disease, there are nearly 430 thousandLGG patients worldwide. Assuming that LGG patients with probabilityof mortality ≥ 0.4 should be clinically intervened, APOLLO couldhelp reduce 240 thousand (55.4%) and 140 thousand (32.4%) unnecessaryinterventions for 36-month and 60-month survival, respectively.
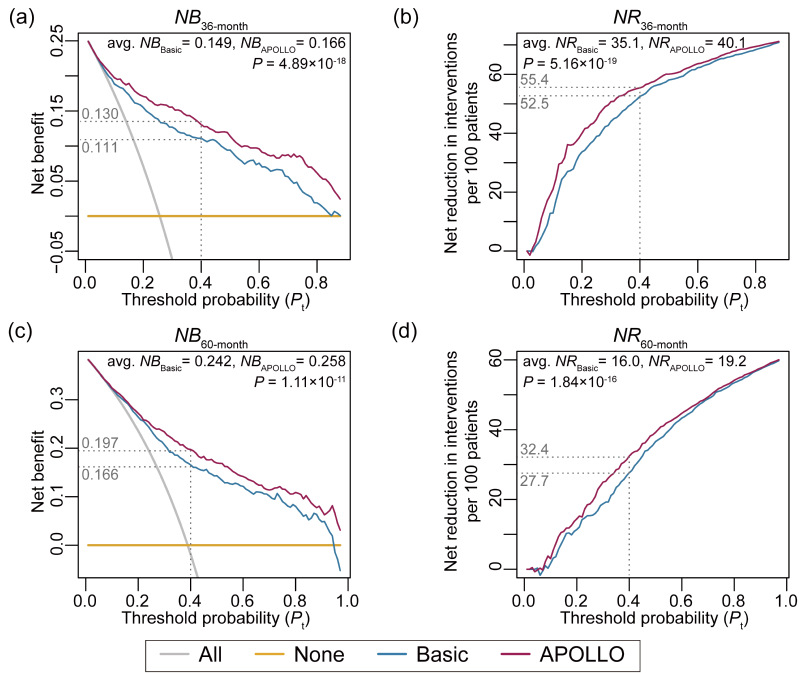
Figure4. Decision curve analysis for clinical application of APOLLO
Tofacilitate the clinical application, Prof. Chen’s Lab alsodeveloped an online platform (Figure5),which can be accessed at http://bigdata.njmu.edu.cn/APOLLO/.
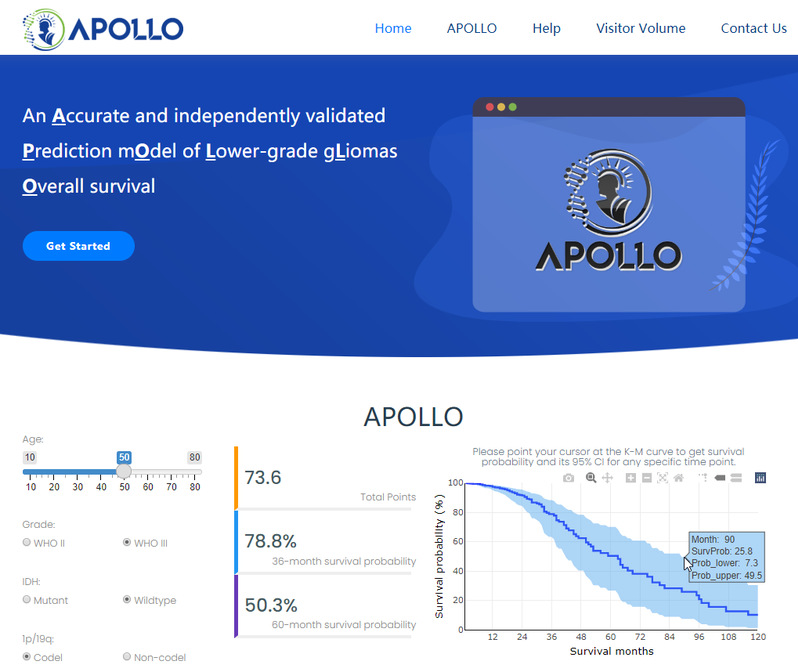
Figure5. APOLLO Online Application Platform
Thisproject was supported by the National Key Research and DevelopmentProgram of China, the National Natural Science Foundation of China,and the NIH of the US RO1.
PhDstudent Jiajin Chen from the Departmentof Biostatistics, Nanjing Medical University; Dr. Sipeng Shen fromtheDepartmentof Biostatistics, Nanjing Medical University; Professor Yi Li fromtheDepartmentof Biostatistics, University of Michigan; and Master student JuanjuanFan from Department of Biostatistics, Nanjing Medical Universityshared the co-first author. Associate Professor Ruyang Zhang andProfessor Feng Chen, from the Department of Biostatistics, School ofPublic Health, Nanjing Medical University, and Professor David C.Christiani from Harvard T.H. Chan School of Public Health are theco-corresponding authors of the work.
ProfessorFeng Chen's Lab has long-termly paid attentiontothe interaction effect identification and prediction modelconstruction of omics data, proposed a 3-D modeling strategy and hasbeen successfully applied to the prognostic prediction for lungcancer (Zhang R, et al. Chest.2020) and lower-grade gliomas (Chen J, et al. eBioMedicine.2022).
Greatthanks to Associate Professor Yongyue Wei and Professor Yang Zhaofrom Department of Biostatistics, Nanjing Medical University,Professor Zhonghua Liu from Department of Statistics and ActuarialScience, University of Hong Kong, Professor Xu Qian from theDepartment of Nutrition, Nanjing Medical University, andundergraduate student Shiyu Xiong and Jingtong Xu from the FirstClinical Medical College, Nanjing Medical University, Master studentChenxu Zhu and PhD student Lijuan Lin from Department ofBiostatistics, Nanjing Medical University, Dr. Xuesi Dong fromNational Cancer Center/National Clinical Research Center, Dr. WeiweiDuan from Department of Bioinformatics, Nanjing Medical University,and also the Chinese Glioma Genome Atlas (CGGA).
Time:April 2022
Publication:eBioMedicine
Title:APOLLO: An accurate and independently validated prediction model oflower-grade gliomas overall survival and a comparative study of modelperformance
Link:https://www.thelancet.com/journals/ebiom/article/PIIS2352-3964(22)00191-8/fulltext
Platform:http://bigdata.njmu.edu.cn/APOLLO/(Computer terminal only)

